Article Published: Genetic capitalism and stabilizing selection of antimicrobial resistance genotypes in Escherichia coli
|
|
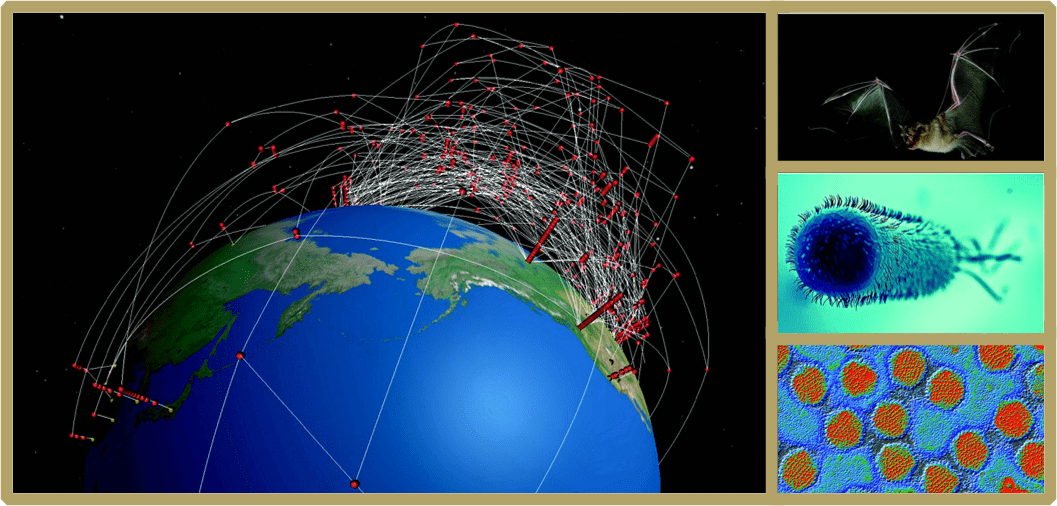 |
|
Cover Image: Global molecular phylogeny of Escherichia coli isolates from the Pathogen Detection project akin to the data used in Ford et al., in this issue of Cladistics entitled “Genetic capitalism and stabilizing selection of antimicrobial resistance genotypes in Escherichia coli”. (Image Credit Daniel Janies and Zachary Witter, University of North Carolina at Charlotte). Small 1: Colored transmission electron micrograph (TEM) of adenovirus particles akin to the data used in Kang et al., entitled “Genomics-based re-examination of the taxonomy and phylogeny of human and simian Mastadenoviruses: An evolving whole genomes approach, revealing putative zoonosis, anthroponosis, and amphizoonosis”. stock photo from GettyImages. Small 2: Escherichia coli. stock photo from GettyImages. Small 3: The Greater horseshoe bat Rhinolophus ferrumequinum. The systematic evidence of bats in this genus and others as reservoirs and origins of current and past viral pathogens is discussed in separate forums by Wenzel as well as by Allard and Brown (stock photo from GettyImages). |
Genetic capitalism and stabilizing selection of antimicrobial resistance genotypes in Escherichia coli
Colby T. Ford, Gabriel Lopez Zenarosa, Kevin B. Smith, David C. Brown, John Williams, Daniel Janies
Cladistics – Volume 36, Issue 4, August 2020, Pages 348-357
Abstract:
Antimicrobial resistance (AMR) in pathogenic strains of bacteria, such as Escherichia coli (E. coli ), adversely impacts personal and public health. In this study, we examine competing hypotheses for the evolution of AMR including (i) ‘genetic capitalism’ in which genotypes that confer antibiotic resistance are gained and not often lost in lineages, and (ii) ‘stabilizing selection’ in which genotypes that confer antibiotic resistance are gained and lost often. To test these hypotheses, we assembled a dataset that includes annotations for 409 AMR genotypes and a phylogenetic tree based on genome‐wide single nucleotide polymorphisms from 29 255 isolates of E. coli collected over the past 134 years. We used phylogenetic methods to count the times each AMR genotype was gained and lost across the tree and used model‐based clustering of the genotypes with respect to their gain and loss rates. We demonstrate that many genotypes cluster to support the hypothesis for genetic capitalism while a few genotypes cluster to support the hypothesis for stabilizing selection. Comparing the sets of genotypes that fall under each of the hypotheses, we found a statistically significant difference in the breakdown of resistance mechanisms through which the AMR genotypes function. The result that many AMR genotypes cluster under genetic capitalism reflects that strong positive selective forces, primarily induced by human industrialization of antibiotics, outweigh the potential fitness costs to the bacterial lineages for carrying the AMR genotypes. We expect genetic capitalism to further drive bacterial lineages to resist antibiotics. We find that antibiotics that function via replacement and efflux tend to behave under stabilizing selection and thus may be valuable in an antibiotic cycling strategy. Read the full article here: https://doi.org/10.1111/cla.12421
